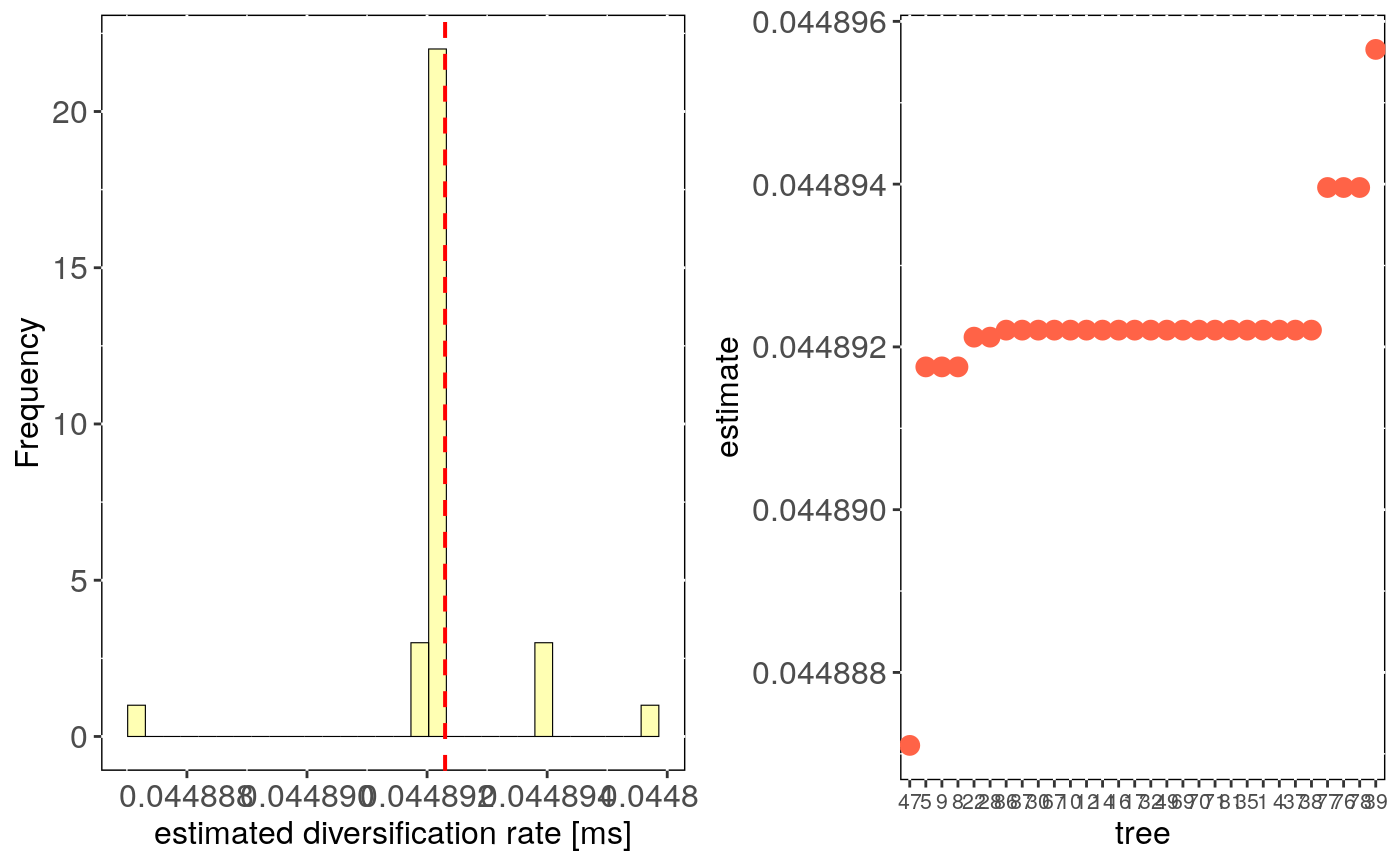
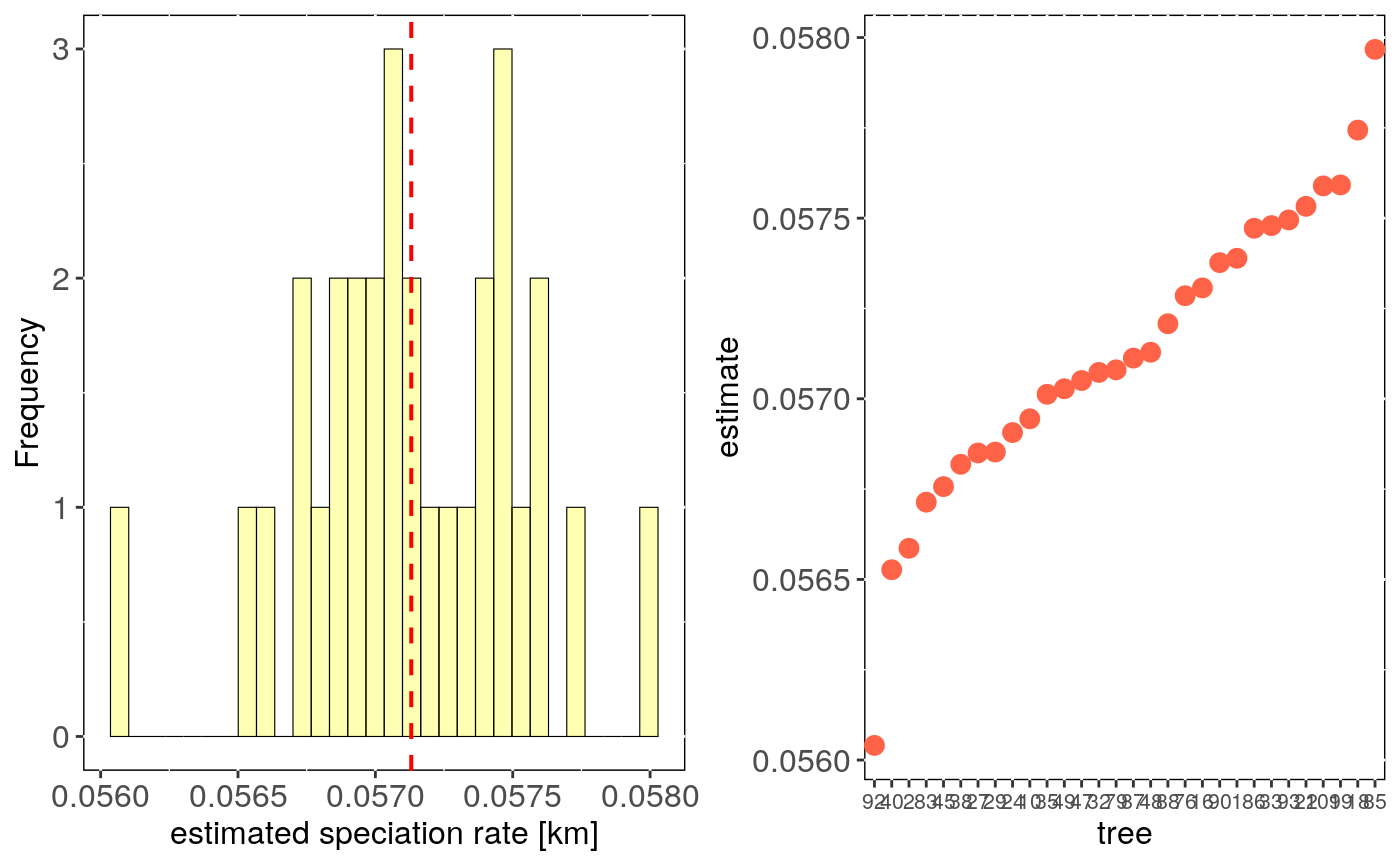
Performs estimates of diversification rate evaluating uncertainty in trees topology.
tree_bd(phy, n.tree = "all", method = "ms", track = F, ...)
Arguments
| phy | A phylogeny (class 'multiPhylo', see ? |
|---|---|
| n.tree | Number of times to repeat the analysis with n different trees picked
randomly in the multiPhylo file. (If |
| method | the method for estimating diversification rate ("ms" or "km") (see Details). |
| track | Print a report tracking function progress (default = TRUE) |
| ... | Further arguments to be passed to |
Value
The function tree_bd returns a list with the following
components:
tree.bd.estimates: Three number, diversification/speciation rate estimate
("Magallon and Sanderson" or "Kendall-Moran") for each run with a different phylogenetic tree.
stats: Main statistics for estimates across trees.CI_low and CI_high are the lower
and upper limits of the 95
Details
This function estimates net diversification rate using bd.ms
(Magallon and Sanderson (2000) method) or speciation rate using bd.km
(Kendall-Moran method) for n trees, randomly picked from a multiPhylo file.
Output can be visualised using sensi_plot.
References
Paterno, G. B., Penone, C. Werner, G. D. A. sensiPhy: An r-package for sensitivity analysis in phylogenetic comparative methods. Methods in Ecology and Evolution 2018, 9(6):1461-1467
Magallon S and MJ Sanderson. 2000. Absolute diversification rates in angiosperm clades. Evolution 55:1762-1780.
See also
Examples
data("primates") # To estimate diversification rate with Magallon and Sanderson method: fit <- tree_bd(phy = primates.phy, n.tree = 30, method = "ms") summary(fit) sensi_plot(fit)# To estimate speciation rate Kendall-Moran method fit <- tree_bd(phy = primates.phy, n.tree = 30, method = "km") summary(fit) sensi_plot(fit)